Table of Contents
Phys gui
All preprocessing is done by
function phys_gui_execute(handles)
Handles are desribed later on in this section Alternatively, you can use a GUI by using the command
phys_gui_working
A figure will pop up which looks something like this:
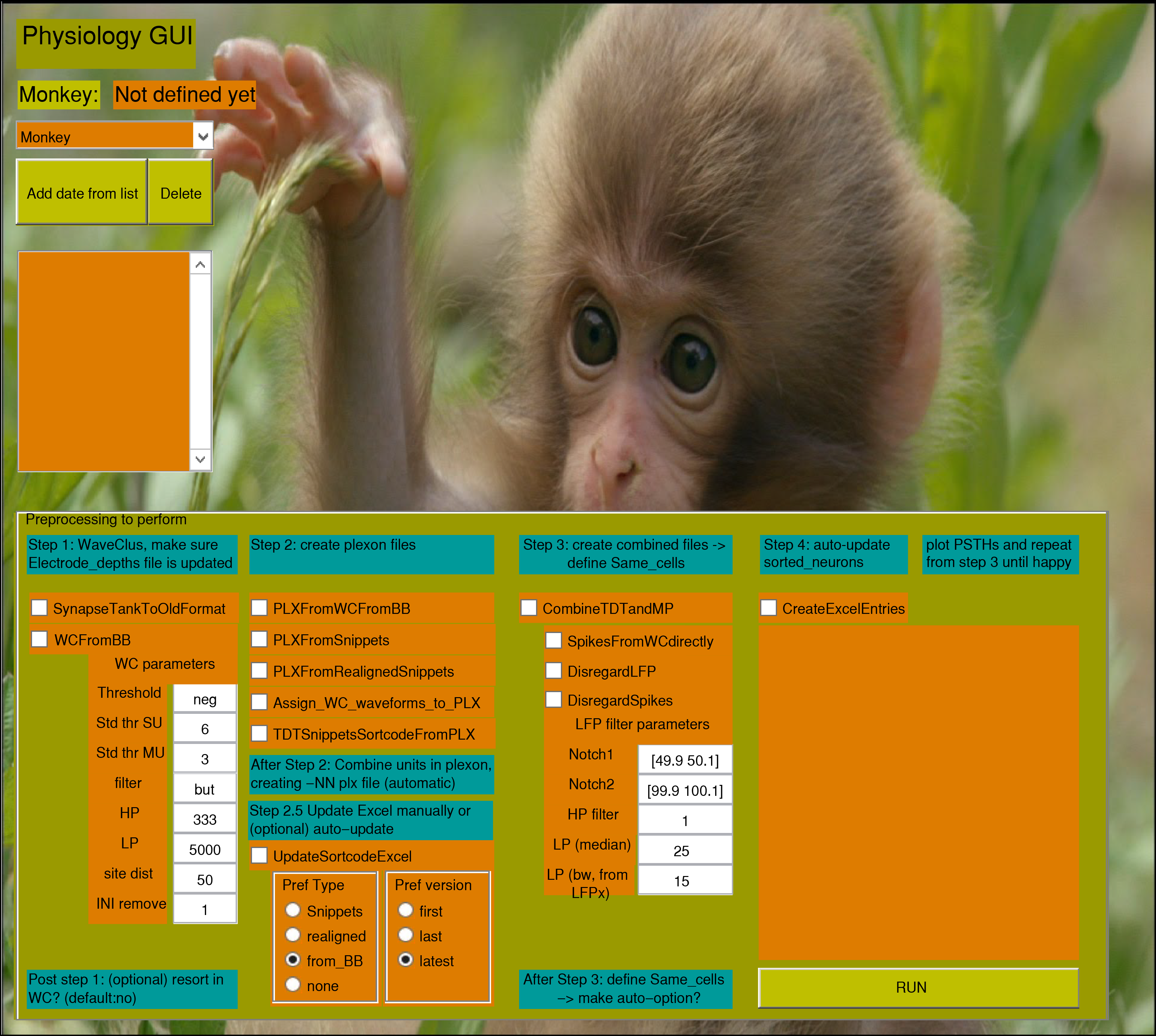 Select your monkey from the dropdown menu
Select your monkey from the dropdown menu
Select the session folders you want to process after hitting the “Add date from list” button
If you hit “Run”, phys_gui will do all the preprocessing that you checked (see below) on the selected folders. The box above will remind you of which steps you selected.
0) Rename folders
In later TDT software versions (synapse - setup 3), the folder naming is different. For converting folders recorded with synapse to the old (our standard) folder naming, check the box “RenameSynapseTankNameToOldTankFormat”.
1) Waveclus pre-clustering
If you want to use the waveclus pipeline, you first need to create waveclus sortcodes. You can do so by checking “WCFromBB”. Make sure, the electrode_depths file is update for the selected blocks, as this is a requirement for the current WC preprocessing pipeline (see …). Folders (WC_Block-N) will be created for each block N, and one additional folder (WC) will contain the clustered data, concatenating for each channel separately all blocks where the electrode was in certain range.
Waveclus pre-clustering settings
| handle | checkbox | Info |
|---|---|---|
| handles.WC.threshold | Threshold | use negative ('neg'), positive ('pos'), or both ('both') |
| handles.WC.StdThrSU | Std thr SU | (higher) threshold for SU detection in std's of data |
| handles.WC.StdThrMU | Std thr MU | (lower) threshold for MU detection in std's of data |
| handles.WC.hp | filter | filter type for HP - median('med') or butterworth('but') |
| handles.WC.hpcutoff | HP | high pass frequency |
| handles.WC.lpcutoff | LP | low pass frequency |
| handles.WC.cell_tracking_distance_limit | site dist | maximum electrode distance for concatenation |
| handles.WC.remove_ini | INI remove | flag to remove ITI before clsutering |
Additional Waveclus settings
For simplicity, some of the potential waveclus settings have been hardcoded inside phys_gui, calling get_WC_settings.
However, when you input the handles directly using phys_gui_execute, you can still define them as you like.
%% defaults handles.RAM = 24; % SYSTEM MEMORY in GB handles.dtyperead = 'single'; % Data TYPE handles.dtypewrite = handles.dtyperead; handles.sys = 'TD'; % RECORDING SYSTEM handles.rawname = '*.tdtch'; % RAW DATAFILES NAME handles.blockfile=0; % not used? % FILTERING: LINE NOISE handles.WC.linenoisecancelation = 0; % 1 for yes; 0 for no handles.WC.linenoisefrequ = 50; % Line noise frequency handles.WC.transform_factor = 0.25; % microVolts per bit for higher accuracy when saved as int16 after filtering; handles.WC.iniartremovel = 1; % ignore first 40 samples % DETECTION handles.WC.w_pre = 10; % N samples for snippet before threshold crossing handles.WC.w_post = 22; % N samples for snipept after threshold crossing handles.WC.ref = 0.001; % 'Refractory period' in seconds handles.WC.int_factor = 1; % for potential interpolation for more datapoints to classify handles.WC.interpolation ='n'; % Interpolationused or not handles.WC.stdmax = 100; % Artifact rejection threshold in std % FEATURE SELECTION handles.WC.features = 'wavpcarawtime'; % features to be considered (in any order: wav(elets),pca,raw (datapoints),time,deriv(ates) handles.WC.wavelet='haar'; % choice of wavelet family for wavelet features handles.WC.exclusioncrit = 'thr'; % this part is weird to me as well, handles.WC.exclusionthr = 0.9; % features are excluded, until no feature pairs are correlated more than exclusionthr %def R^2 = 0.80 handles.WC.maxinputs = 9; % number of feature inputs to the clustering handles.WC.scales = 4; % scales for wavelet decomposition % CLUSTERING handles.WC.num_temp = 18; % number of temperatures handles.WC.mintemp = 0; % minimum temperature handles.WC.maxtemp = 0.18; % maximum temperature handles.WC.tempstep = 0.01; % temperature step handles.WC.SWCycles = 100; % number of montecarlo iterations handles.WC.KNearNeighb = 11; % number of nearest neighbors handles.WC.max_spikes2cluster = 40000; % maximum number of spikes to cluster handles.WC.min_clus_abs = 100; % Minimum cluster size number of spikes handles.WC.min_clus_rel = 0.005; % Minimum cluster size as fraction of all spikes handles.WC.max_nrclasses = 11; % Maximum number of clsuters handles.WC.template_sdnum = 5; % max radius of cluster in std devs. for classifying rest handles.WC.classify_space='features'; % for classifying rest only handles.WC.classify_method= 'linear'; % for classifying rest only % PLOTTING handles.WC.temp_plot = 'log'; % temperature plot in log scale handles.WC.max_spikes2plot = 1000; % maximum number of spikes to plot. handles.WC.max_nrclasses2plot = 8; % not quite sure where this is used
2) Create Plexon files
There are 3 different plexon file types
- If you want (or have) to use the online detected snippets, check “PLXFromSnippets”.
- If you first want to Realign your snippets to the minima, check “PLXFromRealignedSnippets” instead.
- If you want to transfer your waveclus presorting to plexon, check “PLXFromWCFromBB”. In this case you can alternatively skip the plexon step by checking “directly form WC” in the combine TDT and MP step
The two additional options here are
- for taking over WC waveforms to plexon files (keeping the plexon sortcodes) - this was used to counteract a bug where plexon spike resolution in a few cases was terrible
- for creating a TDT sortcode based on plx sorting - this was used in the initial version to then read out snippets with the TDT software
2.5) Create plx file entries
This step can be used to automatically update the “to_use” sheet in the plx file table for the selected monkey and sessions so that the desired (if applicable) plx files will be used for combining ephys and behavior in the next step. Select your preferred sort type, and preferred extension (-01,-02,…) by either picking the first (typically -01), the last, or the latest (last modified)
3) Combine ephys and behavioral data
After you are done with sorting, you can synchronize your recorded data with behavioral data, adding LFP and spike information in the trial structure format that monkeypsych uses. Check “CombineTDTandMP”.
Here you can find the aforementioned option to skip plx files when using waveclus
Another option allows you to disregard spikes (if you are only interested in LFPs). No spike sorting fiels will be needed
There is also an option for disregarding LFP. If you select this option, LFP data in the combined file will not be updated (in case it has been processed previously) or completely left out (if it hasn't been processed before). Use this option as long as you are resorting, it speeds up the processes immensely. You can also add LFP data at a later point.
If you want LFP data to be processed, you can modify the LFP filter settings here:
| handle | checkbox | Info |
|---|---|---|
| handles.LFP.notch_filter1 | Notch1 | first notch (bandstop) filter frequency band |
| handles.LFP.notch_filter2 | Notch2 | second notch (bandstop) filter frequency band |
| handles.LFP.HP_filter | HP filter | butterworth high pass filter frequency |
| handles.LFP.LP_bw_filter | LP(bw, from LFPx) | butterworth filter low pass frequency for pre-filtered LFP (1kHz LFPx) |
| handles.LFP.LP_med_filter | LP(median) | median filter low pass “frequency” N samples window = samplingrate/freq |
4) Create Excel entries
Check “CreateExcelEntries” for creating/updating the automatic sorting sheet of the respective Excel sorting table with the selected data (sessions). Only works after you did all the preceding steps, eventually including filling in the same_cells file.
Running preprocessing without GUI
Although phys_gui is an intuitive tool for and guideline through the preprocessing, you might want to run some more automatized routines. This is completely possible, just use phys_gui_execute with the desired handles as input. To make this easier, simply run it once, the load the newest log file in Y:\Data\All_phys_preprocessing_log. It will contain all the required handles.
